Examples
Digital hologram of a single cell
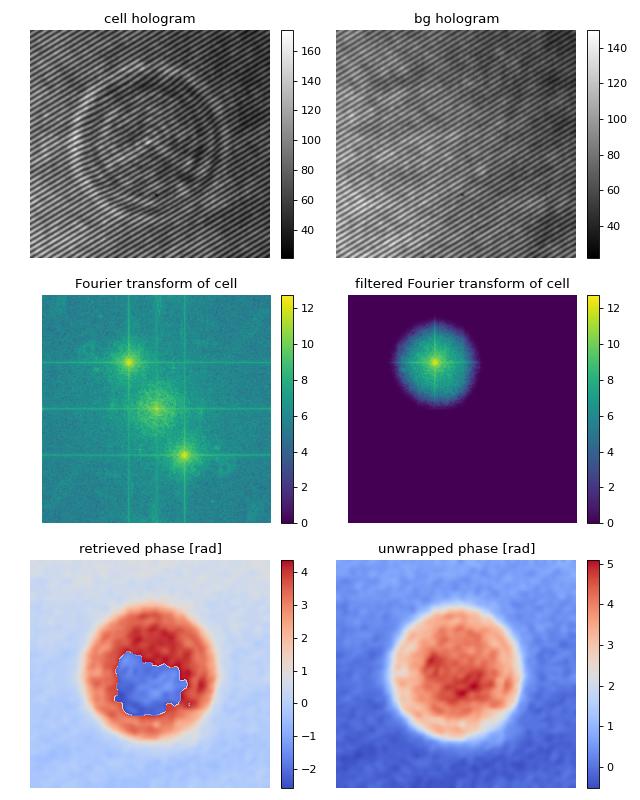
This example illustrates how qpretrieve can be used to analyze digital holograms. The hologram of a single myeloid leukemia cell (HL60) was recorded using off-axis digital holographic microscopy (DHM). Because the phase-retrieval method used in DHM is based on the discrete Fourier transform, there always is a residual background phase tilt which must be removed for further image analysis. The setup used for recording these data is described in reference [SSM+15].
Note that the fringe pattern in this particular example is over-sampled in real space, which is why the sidebands are not properly separated in Fourier space. Thus, the filter in Fourier space is very small which results in a very low effective resolution in the reconstructed phase.
1import matplotlib.pylab as plt
2import numpy as np
3import qpretrieve
4from skimage.restoration import unwrap_phase
5
6# load the experimental data
7edata = np.load("./data/hologram_cell.npz")
8
9holo = qpretrieve.OffAxisHologram(data=edata["data"])
10holo.run_pipeline(
11 # For this hologram, the "smooth disk"
12 # filter yields the best trade-off
13 # between interference from the central
14 # band and image resolution.
15 filter_name="smooth disk",
16 # Set the filter size to half the distance
17 # between the central band and the sideband.
18 filter_size=1/2)
19bg = qpretrieve.OffAxisHologram(data=edata["bg_data"])
20bg.process_like(holo)
21
22phase = holo.phase - bg.phase
23
24# plot the properties of `qpi`
25fig = plt.figure(figsize=(8, 10))
26
27ax1 = plt.subplot(321, title="cell hologram")
28map1 = ax1.imshow(edata["data"], interpolation="bicubic", cmap="gray")
29plt.colorbar(map1, ax=ax1, fraction=.046, pad=0.04)
30
31ax2 = plt.subplot(322, title="bg hologram")
32map2 = ax2.imshow(edata["bg_data"], interpolation="bicubic", cmap="gray")
33plt.colorbar(map2, ax=ax2, fraction=.046, pad=0.04)
34
35ax3 = plt.subplot(323, title="Fourier transform of cell")
36map3 = ax3.imshow(np.log(1 + np.abs(holo.fft_origin)), cmap="viridis")
37plt.colorbar(map3, ax=ax3, fraction=.046, pad=0.04)
38
39ax4 = plt.subplot(324, title="filtered Fourier transform of cell")
40map4 = ax4.imshow(np.log(1 + np.abs(holo.fft_filtered)), cmap="viridis")
41plt.colorbar(map4, ax=ax4, fraction=.046, pad=0.04)
42
43ax5 = plt.subplot(325, title="retrieved phase [rad]")
44map5 = ax5.imshow(phase, cmap="coolwarm")
45plt.colorbar(map5, ax=ax5, fraction=.046, pad=0.04)
46
47ax6 = plt.subplot(326, title="unwrapped phase [rad]")
48map6 = ax6.imshow(unwrap_phase(phase), cmap="coolwarm")
49plt.colorbar(map6, ax=ax6, fraction=.046, pad=0.04)
50
51# disable axes
52[ax.axis("off") for ax in [ax1, ax2, ax3, ax4, ax5, ax6]]
53
54plt.tight_layout()
55plt.show()