Examples
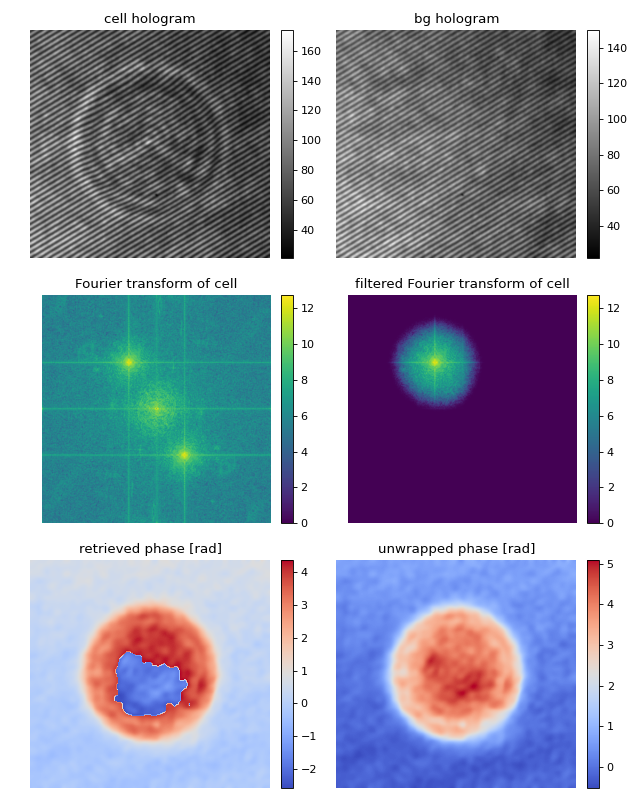
Digital hologram of a single cell
This example illustrates how qpretrieve can be used to analyze digital holograms. The hologram of a single myeloid leukemia cell (HL60) was recorded using off-axis digital holographic microscopy (DHM). Because the phase-retrieval method used in DHM is based on the discrete Fourier transform, there always is a residual background phase tilt which must be removed for further image analysis. The setup used for recording these data is described in reference [SSM+15].
Note that the fringe pattern in this particular example is over-sampled in real space, which is why the sidebands are not properly separated in Fourier space. Thus, the filter in Fourier space is very small which results in a very low effective resolution in the reconstructed phase.
1import matplotlib.pylab as plt
2import numpy as np
3import qpretrieve
4from skimage.restoration import unwrap_phase
5
6# load the experimental data
7edata = np.load("./data/hologram_cell.npz")
8
9holo = qpretrieve.OffAxisHologram(data=edata["data"])
10holo.run_pipeline(
11 # For this hologram, the "smooth disk"
12 # filter yields the best trade-off
13 # between interference from the central
14 # band and image resolution.
15 filter_name="smooth disk",
16 # Set the filter size to half the distance
17 # between the central band and the sideband.
18 filter_size=1/2)
19bg = qpretrieve.OffAxisHologram(data=edata["bg_data"])
20bg.process_like(holo)
21
22phase = holo.phase - bg.phase
23
24# plot the intermediate steps of the analysis pipeline
25fig = plt.figure(figsize=(8, 10))
26
27ax1 = plt.subplot(321, title="cell hologram")
28map1 = ax1.imshow(edata["data"], interpolation="bicubic", cmap="gray")
29plt.colorbar(map1, ax=ax1, fraction=.046, pad=0.04)
30
31ax2 = plt.subplot(322, title="bg hologram")
32map2 = ax2.imshow(edata["bg_data"], interpolation="bicubic", cmap="gray")
33plt.colorbar(map2, ax=ax2, fraction=.046, pad=0.04)
34
35ax3 = plt.subplot(323, title="Fourier transform of cell")
36map3 = ax3.imshow(np.log(1 + np.abs(holo.fft_origin)), cmap="viridis")
37plt.colorbar(map3, ax=ax3, fraction=.046, pad=0.04)
38
39ax4 = plt.subplot(324, title="filtered Fourier transform of cell")
40map4 = ax4.imshow(np.log(1 + np.abs(holo.fft_filtered)), cmap="viridis")
41plt.colorbar(map4, ax=ax4, fraction=.046, pad=0.04)
42
43ax5 = plt.subplot(325, title="retrieved phase [rad]")
44map5 = ax5.imshow(phase, cmap="coolwarm")
45plt.colorbar(map5, ax=ax5, fraction=.046, pad=0.04)
46
47ax6 = plt.subplot(326, title="unwrapped phase [rad]")
48map6 = ax6.imshow(unwrap_phase(phase), cmap="coolwarm")
49plt.colorbar(map6, ax=ax6, fraction=.046, pad=0.04)
50
51# disable axes
52[ax.axis("off") for ax in [ax1, ax2, ax3, ax4, ax5, ax6]]
53
54plt.tight_layout()
55plt.show()
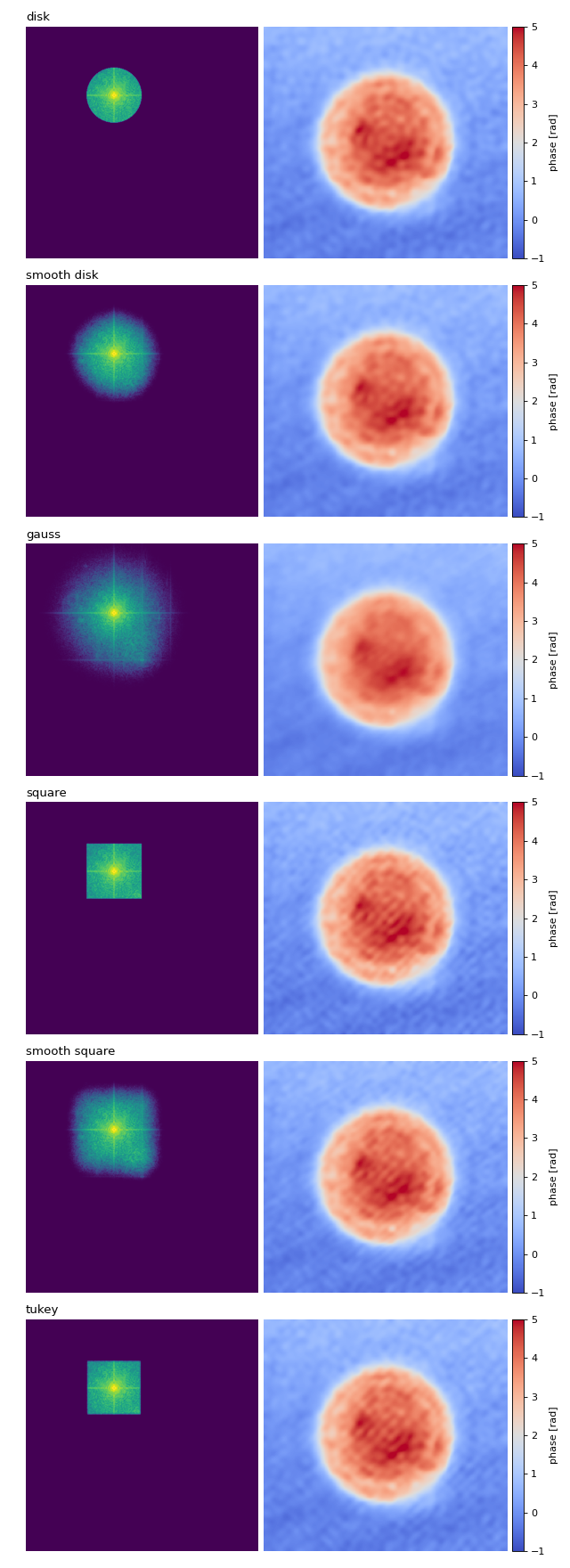
Filter visualization
This example visualizes the different Fourier filtering masks available in qpretrieve.
1import matplotlib.pylab as plt
2import numpy as np
3import qpretrieve
4from skimage.restoration import unwrap_phase
5
6# load the experimental data
7edata = np.load("./data/hologram_cell.npz")
8
9prange = (-1, 5)
10frange = (0, 12)
11
12results = {}
13
14for fn in qpretrieve.filter.available_filters:
15 holo = qpretrieve.OffAxisHologram(data=edata["data"])
16 holo.run_pipeline(
17 filter_name=fn,
18 # Set the filter size to half the distance
19 # between the central band and the sideband.
20 filter_size=1/2)
21 bg = qpretrieve.OffAxisHologram(data=edata["bg_data"])
22 bg.process_like(holo)
23 phase = unwrap_phase(holo.phase - bg.phase)
24 mask = np.log(1 + np.abs(holo.fft_filtered))
25 results[fn] = mask, phase
26
27num_filters = len(results)
28
29# plot the properties of `qpi`
30fig = plt.figure(figsize=(8, 22))
31
32for row, name in enumerate(results):
33 ax1 = plt.subplot(num_filters, 2, 2*row+1)
34 ax1.set_title(name, loc="left")
35 ax1.imshow(results[name][0], vmin=frange[0], vmax=frange[1])
36
37 ax2 = plt.subplot(num_filters, 2, 2*row+2)
38 map2 = ax2.imshow(results[name][1], cmap="coolwarm",
39 vmin=prange[0], vmax=prange[1])
40 plt.colorbar(map2, ax=ax2, fraction=.046, pad=0.02, label="phase [rad]")
41
42 ax1.axis("off")
43 ax2.axis("off")
44
45plt.tight_layout()
46plt.show()
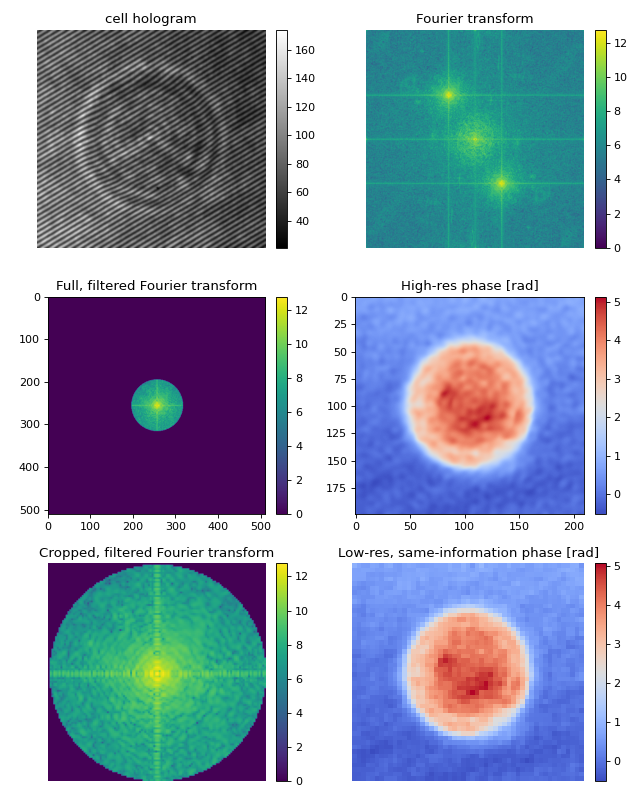
Adequate resolution-scaling via cropping in Fourier space
Applying filters in Fourier space usually means setting a larger fraction of the data in Fourier space to zero. This can considerably slow down your analysis, since the inverse Fouier transform is taking into account a lot of unused frequencies.
By cropping the Fourier domain, the inverse Fourier transform will be faster. The result is an image with fewer pixels (lower resolution), that contains the same information as the unscaled (not cropped in Fourier space) image. There are possibly two disadvantages of performing this cropping operation:
The resolution of the output image (pixel size) is not the same as that of the input (interference) image. Thus, you will have to adapt your colocalization scheme.
If you are using a filter with a non-binary mask (e.g. gauss), then you will lose (little) information when cropping in Fourier space.
What happens when you set filter_name=”smooth disk”? Cropping with scale_to_filter=True is too narrow, because now there are higher frequencies contributing to the final image. To include them, you can set scale_to_filter to a float, e.g. scale_to_filter=1.2.
1import matplotlib.pylab as plt
2import numpy as np
3import qpretrieve
4from skimage.restoration import unwrap_phase
5
6# load the experimental data
7edata = np.load("./data/hologram_cell.npz")
8
9results = []
10
11holo = qpretrieve.OffAxisHologram(data=edata["data"])
12bg = qpretrieve.OffAxisHologram(data=edata["bg_data"])
13ft_orig = np.log(1 + np.abs(holo.fft_origin), dtype=float)
14
15holo.run_pipeline(filter_name="disk", filter_size=1/2,
16 scale_to_filter=False)
17bg.process_like(holo)
18phase_highres = unwrap_phase(holo.phase - bg.phase)
19ft_highres = np.log(1 + np.abs(holo.fft.fft_used), dtype=float)
20
21holo.run_pipeline(filter_name="disk", filter_size=1/2,
22 scale_to_filter=True)
23bg.process_like(holo)
24phase_scalerad = unwrap_phase(holo.phase - bg.phase)
25ft_scalerad = np.log(1 + np.abs(holo.fft.fft_used), dtype=float)
26
27# plot the intermediate steps of the analysis pipeline
28fig = plt.figure(figsize=(8, 10))
29
30ax1 = plt.subplot(321, title="cell hologram")
31map1 = ax1.imshow(edata["data"], interpolation="bicubic", cmap="gray")
32plt.colorbar(map1, ax=ax1, fraction=.046, pad=0.04)
33
34ax2 = plt.subplot(322, title="Fourier transform")
35map2 = ax2.imshow(ft_orig, cmap="viridis")
36plt.colorbar(map2, ax=ax2, fraction=.046, pad=0.04)
37
38ax3 = plt.subplot(323, title="Full, filtered Fourier transform")
39map3 = ax3.imshow(ft_highres, cmap="viridis")
40plt.colorbar(map3, ax=ax3, fraction=.046, pad=0.04)
41
42ax4 = plt.subplot(324, title="High-res phase [rad]")
43map4 = ax4.imshow(phase_highres, cmap="coolwarm")
44plt.colorbar(map4, ax=ax4, fraction=.046, pad=0.04)
45
46ax5 = plt.subplot(325, title="Cropped, filtered Fourier transform")
47map5 = ax5.imshow(ft_scalerad, cmap="viridis")
48plt.colorbar(map5, ax=ax5, fraction=.046, pad=0.04)
49
50ax6 = plt.subplot(326, title="Low-res, same-information phase [rad]")
51map6 = ax6.imshow(phase_scalerad, cmap="coolwarm")
52plt.colorbar(map6, ax=ax6, fraction=.046, pad=0.04)
53
54# disable axes
55[ax.axis("off") for ax in [ax1, ax2, ax5, ax6]]
56
57plt.tight_layout()
58plt.show()